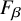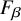Highlights
-
1. What is already known
-
• Biomedical concept normalization is the mapping of free-text phrases (example: “Type 2 Diabetes Mellitus”) to a standardized coded representation (example: UMLS “C0011860”).
-
• A number of commonly used tools and methods for biomedical concept normalization exist. Such tools tend to achieve modestly high recall and poor precision, though performance varies depending on data and tools.
-
• Large Language Models (LLMs) may perform well for certain reasoning and Natural Language Processing (NLP) tasks. Current evidence suggests LLMs used alone tend to perform poorly for normalization, however.
-
-
2. What is new
-
• We contribute an empirical evaluation of the use of commonly used biomedical concept normalization tools and methods together with LLMs in a multi-step process.
-
• Using a reasonably large publicly available biomedical dataset, 2 LLMs, and 4 normalization methods, we demonstrate that our multi-step approach can improve F1 scores between +7.3% and +21.7%.
-
-
3. Potential Impact for Research Synthesis Methods readers
-
• The need for normalization is common in many biomedical tasks and analyses. Readers can utilize our findings in understanding possible tradeoffs in precision, recall, the use of open- vs. closed-source LLMs, and associated potential time and cost in their decision-making.
-
1 Introduction
Biomedical entity normalization, also known as entity linking or grounding, is the process of mapping spans of text, such as conditions, procedures, or medications, into coded representations, such as Unified Medical Language System (UMLS) codes. Normalization is critical to biomedical research because the richness of free-text data, such as information within progress notes, can often be fully leveraged only after mapping words and phrases into structured, coded representations suitable for analysis. For example, identifying patients with Type 2 Diabetes Mellitus using free-text narratives could be challenging with keyword search alone, given the variety of possible phrasings (“T2DM,” “Hyperglycemia,” “Glucose intolerance,” etc.) Searching instead for a normalized representation (e.g., UMLS “C0011860”) simplifies this process while greatly improving recall. Coded terms can subsequently be linked to related terms within ontologies and so on. Recent state-of-the-art Large Language Models (LLMs), in turn, have shown great potential and high performance in a variety of biomedical natural language processing (NLP) tasks,Reference Xu, Chen and Peng 1 – Reference Dobbins, Han and Zhou 6 but their application for normalization have focused on prediction of comparatively smaller vocabularies,Reference Wang, Liu, Yang and Weng 7 , Reference Soroush, Glicksberg and Zimlichman 8 rephrasing and text cleaning without pruning,Reference Abdulnazar, Roller, Schulz and Kreuzthaler 9 or used datasets whose generalizability to scientific and clinical documents is unclear.Reference Chen, Li, Ding and Cleveland 10 Moreover, while biomedical informaticians and researchers often leverage rule-based systems, such as MetaMapLiteReference Demner-Fushman, Rogers and Aronson 11 or cTAKESReference Savova, Masanz and Ogren 12 for normalization, few studies have evaluated the use of LLMs working in concert with existing normalization systems.Reference Abdulnazar, Roller, Schulz and Kreuzthaler 9 , Reference Borchert, Llorca and Schapranow 13
In this study, we evaluate the use of two widely used LLMs, one closed-source (GPT-3.5-turbo) 14 and one open (Vicuna-13b,Reference Chiang, Li and Lin 15 a fine-tuned variation of LlamaReference Touvron, Lavril and Izacard 16 ), alongside widely used normalization techniques and libraries within the informatics community, on a large human-annotated corpus of biomedical abstracts and UMLS concepts.Reference Mohan and Li 17 We aim to contribute to improving performance for a common scenario within biomedical informatics research: the need to extract normalized concepts from a large corpus of documents, where fine-tuning a domain-specific model for the task is not practical or possible (for example due to the lack of a gold standard annotation or time). In such cases, many researchers use applications such as MetaMapLite,Reference Demner-Fushman, Rogers and Aronson 11 QuickUMLS,Reference Soldaini and Goharian 18 cTAKES,Reference Savova, Masanz and Ogren 12 CLAMP,Reference Soysal, Wang and Jiang 19 which tend to be rule-based and relatively fast and scalable in concept extraction, but often with only moderate recall and relatively low precision. Alternatively, researchers may also utilize word embeddings,Reference Kalyan and Sangeetha 20 dense vector representations of phrases or documents which can be compared to similar embeddings of UMLS or other concepts, then normalized by selecting the closest concepts as measured by cosine similarity, for example. We seek to explore the question: How can comparatively small, widely available LLMs be leveraged to improve upon these baseline biomedical concept normalization performance? We examine a variety of prompting strategies with a focus on two specific areas where LLMs may aid improvement alongside existing normalization systems. All code used in this study is available at https://github.com/ndobb/llm-normalization/ and https://zenodo.org/records/14715161.
2 Background and significance
The process of concept normalization from biomedical free-text documents has been studied extensively, and a number of widely used normalization systems exist. Such systems tend to be rule-based, parsing text and matching based on lexical and syntactic heuristics,Reference Demner-Fushman, Rogers and Aronson 11 , Reference Savova, Masanz and Ogren 12 , Reference Soldaini and Goharian 18 , Reference Aronson and Lang 21 though more recent systems, such as the CLAMP toolkit,Reference Soysal, Wang and Jiang 19 also incorporate machine learning based models for sentence boundary detection and named entity recognition.
Related to normalization, Narayan et al. Reference Narayan, Chami, Orr, Arora and Ré 22 explored the use of LLMs using OpenAI’s GPT-3Reference Mann, Ryder and Subbiah 23 on publicly available datasets to evaluate entity matching between datasets (e.g., determining if two products are the same based on their descriptions), error detection, and data imputation. Peeters et al. Reference Peeters and Bizer 24 , Reference Peeters and Bizer 25 similarly expanded this line of research using GPT-3.5 and systematically explored various prompting strategies. Other work in this area has been driven by ontology researchers aiming to determine how LLMs may be leveraged for entity alignment and relation prediction (e.g,. is-a, subsumes) to automate ontology generation and error checking.Reference Babaei Giglou, D’Souza and Auer 2 , Reference Hertling and Paulheim 26 – Reference Matentzoglu, Caufield and Hegde 28
Within the health domain, Yang et al. Reference Yang, Marrese-Taylor, Ke, Cheng, Chen and Li 29 used a Retrieval Augmented Generation (RAG) based approachReference Lewis, Perez and Piktus 30 to inject UMLS-derived context within prompts to improve question-answering performance. Specific to normalization, Borchert et al. used an LLM-based pipeline to transform phrases into simplified phrases which were subsequently normalized to coded representations using term frequency-inverse document frequency (TF-IDF). Soroush et al. Reference Soroush, Glicksberg and Zimlichman 8 analyzed the use of GPT models for generating ICD-9, ICD-10, and CPT codes from text-descriptions, finding both GPT-3.5-turbo and GPT-4 to perform relatively poorly, with accuracy under 50%. Soroush et al.’s findings suggests that LLMs alone may be inappropriate tools for concept normalization. This challenge may be even more so for UMLS concepts (as opposed to ICD-10), which include a far larger number of longer, total codes which themselves carry no inherent meaning and imply no relation to other codes (for example, the sequence of characters for the UMLS code “C0011860” are essentially arbitrary, unlike codes which begin with “E” in ICD-10, which relate in some fashion to diabetes mellitus). Wang et al. Reference Wang, Liu, Yang and Weng 7 fine-tuned a Llama 2 to text to normalize Human Phenotype (HPO) codes and demonstrated better performance than GPT-3.5-turbo, though the vocabulary (roughly 17,000 codes) was notably smaller than the entirety of the UMLS and it is unclear how well their methods would generalize to larger vocabularies. Using embeddings, Tutubaline et al. Reference Tutubalina, Miftahutdinov, Nikolenko and Malykh 31 and Belousov et al. Reference Belousov, Dixon and Nenadic 32 evaluated the use of embeddings and various neural architectures using health-related posts from social media, achieving accuracies of 70% and 88%, respectively.
2.1 Key contributions
As current state-of-the-art LLMs alone tend to perform poorly at normalization, we seek to explore how well they can improve baseline normalization performance when used alongside and augmenting widely-used existing normalization software. To do so, we:
-
1. Establish baseline precision, recall, F1 and
 $F_{\beta }$
performance using several common normalization applications and embeddings on a large set of human-annotated, normalized condition mentions.
$F_{\beta }$
performance using several common normalization applications and embeddings on a large set of human-annotated, normalized condition mentions. -
2. Use both open- and closed-source LLMs for synonym and alternative phrasing generation, which we further normalize in order to maximize recall.
-
3. Explore prompting strategies for normalized concept pruning to subsequently balance precision.
-
4. Utilize combinations of normalization systems and LLMs to robustly evaluate end-to-end normalization strategies aimed at maximizing
 $F_{\beta }$
, a harmonic mean of precision and recall weighted toward recall.
$F_{\beta }$
, a harmonic mean of precision and recall weighted toward recall.
We designed our experiments with a focus on practicality and scale, intended to inform biomedical research efforts requiring normalization of large-scale unannotated clinical text repositories while minimizing potential cost. Our results can be leveraged by researchers with various goals in mind, such maximizing recall or precision, or in understanding cost and performance strategies in various prompting strategies.
3 Materials and methods
3.1 Language models
Biomedical concept normalization efforts often involve thousands or even millions of documents.Reference Xu, Musen and Shah 33 , Reference Wu, Liu and Li 34 In such projects the need for normalization processes at speed and without significant cost (e.g., less than 1 second and under $0.01 per document) are thus necessary. We chose two language models, one publicly available and one proprietary, as we believed they achieved a reasonable tradeoff between strong (though not state-of-the-art)Reference Li, Gupta, Bhaduri, Sathiadoss, Bhatnagar and Chong 35 – Reference Sun, Hadjiiski and Gormley 38 performance, inference speed, and cost.
-
1. GPT-3.5-turbo is the primary model behind ChatGPT 14 at the time of this writing. GPT-3.5-turbo has been demonstrated to perform well on a variety of tasks within the clinical domain,Reference Liu, Wang and Liu 39 , Reference Rao, Pang and Kim 40 including authoring letters to patients,Reference Ali, Dobbs, Hutchings and Whitaker 41 decision support,Reference Liu, Wright and Patterson 42 medical question-answering,Reference Johnson, Goodman and Patrinely 43 interpreting radiology reports,Reference Jeblick, Schachtner and Dexl 44 and various clinical NLP tasks.Reference Li, Dada, Puladi, Kleesiek and Egger 45 – Reference Hu, Chen and Du 47 We used the gpt-3.5-turbo-0125 model within OpenAI for our experiments.
-
2. Vicuna is an open-source model fine-tuned from the original LlaMA modelReference Touvron, Lavril and Izacard 16 using user-shared responses from ChatGTP.Reference Chiang, Li and Lin 15 As Vicuna was thus tuned to respond similarly to ChatGPT but is freely available and also comparatively smaller, we chose Vicuna as a reasonable alternative capable of running locally without the need for significant infrastructure. We used the quantized vicuna-13b-v1.5.Q4_K_M.gguf modelFootnote i for our experiments.
3.2 Normalization systems
We chose four methods and software libraries as our baseline normalization systems:
-
1. OpenAI Embeddings are dense vector representations of source text phrases or passages that enable semantic similarity search. While embeddings may be generated by a variety of methods, including open-source, we utilize OpenAI-generated UMLS and named entity embeddings as these have been shown to perform well for search tasks,Reference Xian, Teofili, Pradeep and Lin 48 can be generated relatively quickly and at scale, and are widely used at the time of this writing.
-
2. MetaMapLite Reference Demner-Fushman, Rogers and Aronson 11 is a Java-based implementation of the original MetaMap algorithm.Reference Aronson and Lang 21 MetaMapLite uses a Lucene-basedReference Białecki, Muir, Ingersoll and Imagination 49 dictionary lookup approach for normalizing concepts indexed by UMLS concept source text, abbreviations, and source term types.
-
3. QuickUMLS Reference Soldaini and Goharian 18 is a lightweight Python-based implementation of the CPMerge algorithm.Reference Okazaki and Tsujii 50 QuickUMLS is designed to achieve similar recall and precision to MetaMap and cTAKES but much faster.
-
4. BM25, or more formally Okapi BM25, is a widely used ranking and retrieval algorithm.Reference Whissell and Clarke 51 We used a Python implementation of BM25 from the retriv library.Footnote ii We indexed BM25 using the preferred terms of UMLS concepts related to diseases and conditions.
3.3 Dataset
MedMentionsReference Mohan and Li 17 is a human-annotated dataset of over 4,000 biomedical abstracts. Each abstract contains mentions of biomedical concept terms and their most appropriate corresponding UMLS concept ID. We chose MedMentions because of both the high-quality of the annotations and the large relative size of the corpus.
As the focus of this study is to inform low-resource scenarios where researchers are unable to train or fine-tune models for normalization, we utilized only the test set of MedMentions “full” version for evaluation purposes. The test set consists of 839 abstracts with 70,405 UMLS concepts. As we aimed to evaluate our LLM-augmented normalization methods rather than exhaustively validate the corpus using the entirety of the UMLS, we limited our experiments to 5,523 UMLS concept annotations related to disease and conditions by using the UMLS semantic types “TUI” (Type Unique Identifier).
3.4 Normalization strategy
We aimed to leverage the LLMs described to improve the baseline normalization performance of MetaMapLite, QuickUMLS, and BM25 in a relatively simple pipeline-like process. Importantly, we assume a scenario where named entities within a given corpus are already known, but not normalized. In other words, we imagine a case where a named entity recognition (NER) algorithm has already identified candidate token spans within a corpus (for example, “cystic fibrosis” at character indices 250-265 in document 1) but UMLS concepts are not known. We seek to augment traditional normalization processes with the following strategies:
-
1. Synonym and Alternate Phrasing Generation - We prompt an LLM to generate alternative phrasings or synonyms of a given input text span to be normalized. We then use our normalization systems to attempt to normalize both the source text span, as well as candidate alternative spans. This step seeks to maximize recall.
-
2. Candidate Concept Pruning - After normalization, we again prompt an LLM to filter out inappropriate concepts, using both the preferred term and semantic type of a given candidate concept as well as surrounding text context of the original identified span. This step seeks to improve precision.
Figure 1 shows a visual example of this process.

Figure 1 Diagram of our multi-stage normalization strategy.
3.5 Evaluation
Our evaluation process was as follow:
-
1. Using all three normalization systems, we established baseline normalization results by individually executing each on all condition concepts in the test set of the MedMentions corpus.
-
2. We prompted both LLMs to generate candidate alternative phrasings and synonyms for all test set concepts, then used our normalization systems to subsequently normalize the additional concepts as well, focusing on recall.
-
3. Next, we sought to identify an optimal prompting strategy for concept pruning. To do so, we experimented using a random sample of 1,000 test set concepts and the Vicuna model with various prompt structures. These were:
-
(a) Multiple Choice - We included all candidate concepts in a single prompt, instructing the LLM to output a list of appropriate concepts. Within the multiple choice approach, we further experimented with outputting UMLS concept concept IDs (CUIs) versus indices in which they were presented (e.g., “C0010674” vs. “2”). We hypothesized that simpler index-based output may may perform slightly better than CUIs. We additionally evaluated chain-of-thought prompting, instructing the model to “think step-by-step” and provide reasoning for a given output.
-
(b) Binary Choice - We evaluated each candidate concept independently, with an additional LLM prompt and response for each concept. We hypothesized that a simpler prompt and question may also lead to better performance, though at the cost of far more overall prompts needed.
Figure 2 shows examples of these strategies.

Figure 2 Visual example of our Multiple Choice and Binary Choice prompting strategies.
In addition, for each prompt strategy combination, we also experimented with a post-processing step to always accept the first candidate concept, even if rejected in the concept pruning phase. If normalized candidates were returned from the original utterance, we use the first of these, which are typically sorted by closest syntactic match. If no normalized candidates were returned from the original utterance but were returned from synonyms, we accept the first of these. We refer to this strategy hereafter as Top1.
For scoring purposes, we make the explicit assumption that recall is ultimately more important than precision in this task. For example, many downstream analyses of normalized, extracted data often look for specific CUI values (e.g., identifying patients with heart failure “C0018801” as clinical trial candidates). Thus false positive CUIs may be ignored in such cases with relatively low harm. We therefore consider
 $F_{\beta }$
as our metric to optimize, with recall having greater weight than precision (
$F_{\beta }$
as our metric to optimize, with recall having greater weight than precision (
 $\beta = 2$
):
$\beta = 2$
):  $$\begin{align*}F_{\beta} = (1 + \beta^2) * \frac{precision*recall}{(\beta^2*precision)+recall}. \end{align*}$$
$$\begin{align*}F_{\beta} = (1 + \beta^2) * \frac{precision*recall}{(\beta^2*precision)+recall}. \end{align*}$$
For context we also provide the traditional F1 harmonic mean of precision and recall:
 $$\begin{align*}F1 = 2*\frac{precision*recall}{precision+recall}. \end{align*}$$
$$\begin{align*}F1 = 2*\frac{precision*recall}{precision+recall}. \end{align*}$$
-
4 Results
Table 1 shows results of our baseline and synonym and alternate phrasing phrasing experiments. As our results at this stage include candidate concepts from our LLM-generated alternate phrasings without pruning, the improvements shown are thus the upper bound of recall. We prompted each LLM to return at most 3 alternate phrasings (Supplementary Figure 1). For each alternate phrasing, we accepted only the first normalized concept returned, if any. While both LLMs showed recall improvement over baseline results with each normalization system, GPT-3.5-turbo showed a greater recall improvement for each over Vicuna. With the exception of BM25 and embeddings (which already had reasonably high recalls of 77.7% and 72% at baseline), each normalization system showed an improvement in recall of over 10% for both LLMs.
Table 1 Results of our first experiments using LLMs to generate and normalize synonyms and alternative phrasings of an initial utterance

Note: We aimed to improve recall over baseline results. The highest scores for each metric are shown in bold.
Next, we aimed to determine optimal prompt and scoring strategies for concept pruning, using variants of our prompt strategies. Given the large number of potential LLM responses needed due to various combinations of prompt strategies, we chose to use only the Vicuna LLM and a random subset of 1,000 examples from the test set. Results are shown in Table 2.
Table 2 Results of experiments to determine an optimal prompting strategy for concept pruning

Note: We used a randomly-chosen subset of 1,000 test set concepts and contexts. The highest scores for each metric are shown in bold.
For each normalization system, as the binary prompt strategy with chain-of-thought showed the best performance by
![]() $F_{\beta }$
among our test set sample using LLM-chosen UMLS concepts, we chose to use this in our final experiment. As automatically including the first candidate concept also improved recall (though at the expense of some precision), we also included the “Top1” strategy in our final experiment. Results are shown in Table 3.
$F_{\beta }$
among our test set sample using LLM-chosen UMLS concepts, we chose to use this in our final experiment. As automatically including the first candidate concept also improved recall (though at the expense of some precision), we also included the “Top1” strategy in our final experiment. Results are shown in Table 3.
Table 3 Results of our final experiment to apply our best-performing prompting strategy using all normalization systems and LLMs

Despite its presumably smaller number of parameters than the proprietary GPT-3.5-turbo model, the smaller Vicuna model achieved the highest F1 scores over GPT-3.5-turbo in all experiments. Between the two models,
![]() $F_{\beta }$
scores were generally closer with the exception of BM25, where Vicuna achieved notably better
$F_{\beta }$
scores were generally closer with the exception of BM25, where Vicuna achieved notably better
![]() $F_{\beta }$
than GPT-3.5-turbo (CoT: +3.9 and CoT+Top1: +5.1). Compared to baseline normalization
$F_{\beta }$
than GPT-3.5-turbo (CoT: +3.9 and CoT+Top1: +5.1). Compared to baseline normalization
![]() $F_{\beta }$
results in our first experiment (shown in Table 1), the addition of the Vicuna model for end-to-end alternative phrasing and best pruning strategy led to over 10% improvement in all normalization systems (Embeddings: +20.2, MetaMapLite: +10.8, QuickUMLS: +14.7, BM25: +15.6).
$F_{\beta }$
results in our first experiment (shown in Table 1), the addition of the Vicuna model for end-to-end alternative phrasing and best pruning strategy led to over 10% improvement in all normalization systems (Embeddings: +20.2, MetaMapLite: +10.8, QuickUMLS: +14.7, BM25: +15.6).
Full prompts for all experiments are included in the Supplementary Material.
5 Discussion
Our results demonstrate that existing LLMs can be leveraged to greatly increase performance of widely-used biomedical normalization systems without fine-tuning. Our two-step process of leveraging LLMs for alternate phrasing generation and subsequent concept pruning demonstrates improvements to
![]() $F_{\beta }$
(best: +15.6) and F1 (best: +19.5) in each combination LLM and normalization system we experimented with, as well as higher recall in all experiments with the exception of one (BM25 using Vicuna with Binary-Choice+CoT prompting strategy).
$F_{\beta }$
(best: +15.6) and F1 (best: +19.5) in each combination LLM and normalization system we experimented with, as well as higher recall in all experiments with the exception of one (BM25 using Vicuna with Binary-Choice+CoT prompting strategy).
Moreover, we demonstrate that these results can be achieved using non-state-of-the-art, smaller models, both open-source and proprietary. Our results show that the publicly-available Vicuna 13b quantized model can achieve results that surpass GPT-3.5-turbo in
![]() $F_{\beta }$
and F1 under certain circumstances. As these smaller models can respond relatively quickly and at relatively low cost, they also lend themselves to scalability, in concert with normalization systems. Our results also empirically demonstrate that while widely used for various NLP tasks,Reference Kalyan and Sangeetha
20
for biomedical concept normalization the use of embeddings alone may perform substantially worse than our best performing multi-step normalization strategy (embeddings baseline:
$F_{\beta }$
and F1 under certain circumstances. As these smaller models can respond relatively quickly and at relatively low cost, they also lend themselves to scalability, in concert with normalization systems. Our results also empirically demonstrate that while widely used for various NLP tasks,Reference Kalyan and Sangeetha
20
for biomedical concept normalization the use of embeddings alone may perform substantially worse than our best performing multi-step normalization strategy (embeddings baseline:
![]() $F_{\beta }$
40.0%, best performing strategy:
$F_{\beta }$
40.0%, best performing strategy:
![]() $F_{\beta }$
65.6%), though this comes at the expense of significantly more processing per named entity, and further experimentation is needed.
$F_{\beta }$
65.6%), though this comes at the expense of significantly more processing per named entity, and further experimentation is needed.
5.1 Limitations
This study had a number of limitations. First, we used only a single dataset, MedMentions, and only concepts related to diseases and conditions. It is possible that other datasets, textual contexts, domains and concept semantic types (e.g., medications) may show different results. As the MedMentions test set includes over 5,000 concepts from a large variety of biomedical abstracts, however, we argue that the dataset is nonetheless useful and reasonable for establishing the efficacy of our methods. Next, we evaluated only two large language models, and potentially there are a growing list of others that may be included. As we intended to demonstrate that existing, readily-available, smaller models encode sufficient biomedical knowledge while also remaining inexpensive and highly responsive, we argue these models are highly suitable to common research situations where a large corpora of documents require normalization at scale while model fine-tuning is not possible or desirable. Additionally, we assume a situation where named entities within a corpus are already identified but not yet normalized, which may not always be the case. As reasonably well-performing NER models, including open-source, are readily-available, we believe this is a reasonable assumption. Further, we used only a subset of 1,000 test set concepts for prompt evaluation. In theory, though randomly chosen, it is possible that the prompt structure results from our 1,000 concept subset may not generalize to other concepts in the test set. Given the computational and time costs (each concept required approximately 10-20 LLM inferences per model due to the variety of parameters and strategies) as well as the size of the subset used, we believe this is a reasonable tradeoff and the systemic differences between the subset and other concepts (which would invalidate our findings) in Table 2 to be possible but unlikely.
5.2 Future work
In future work, we intend to apply these approaches to existing corpora of other domains (such as progress notes), as well as evaluate cost and scaling factors in greater detail.
6 Conclusion
This study demonstrates that smaller, widely-available large language models can be readily leveraged alongside existing normalization software to achieve greater precision, recall,
![]() $F_{\beta }$
and F1 in clinical documents. We empirically evaluated two language models in combination with four widely-user normalization systems using a variety of prompting and processing strategies. The methods discussed here can be readily adapted and used by research teams.
$F_{\beta }$
and F1 in clinical documents. We empirically evaluated two language models in combination with four widely-user normalization systems using a variety of prompting and processing strategies. The methods discussed here can be readily adapted and used by research teams.
Acknowledgements
We thank Meliha Yetisgen, Fei Xia, Ozlem Uzuner, and Kevin Lybarger for their thoughts and suggestions in the planning phase of this project.
Author contributions
NJD conceived of and executed all experiments and wrote the manuscript.
Competing interest statement
NJD has consulted for TriNetX, LLC, and the National Institute of Health.
Data availability statement
All code and data used in this study are available at https://github.com/ndobb/llm-normalization/ and https://zenodo.org/records/14715161. The MedMentions dataset is available to https://github.com/chanzuckerberg/MedMentions.
Funding statement
This study was supported in part by the National Library of Medicine under Award Number R15LM013209 and by the National Center for Advancing Translational Sciences of National Institutes of Health under Award Number UL1TR002319. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/rsm.2025.9.