Introduction
Stephanofilaria is an economically important genus of nematodes (order: Spirurida, family: Filariidae) that causes ulcerative dermal lesions in several species of large mammals globally. About a dozen species have been morphologically described in this genus (Hodda Reference Hodda2022). These include Stephanofilaria dinniki, which infects the critically endangered black rhinoceros (Round, Reference Round1964); Stephanofilaria thelazioides, which infects the hippopotamuses (Boomker et al. Reference Boomker, Bain, Chabaud and Kriek1995); Stephanofilaria boomkeri, which infects domestic and wild African suids (Bain et al. Reference Bain, Van der Lugt and Kazadi1996); Stephanofilaria zaheeri, which infects water buffalo Bubalus bubalis; and Stephanofilaria srivastavai, which infects the Asian elephant, Elephas maximus (Bhattacharjee Reference Bhattacharjee1967). In addition, several species of Stephanofilaria that parasitize domestic bovids including cattle in different parts of the world have been described. Stephanofilaria stilesi (Chitwood Reference Chitwood1934), has been reported in the USA and Canada, Stephanofilaria sp. in Australia, Stephanofilaria assamensis (Singh Reference Singh1958) in India and Russia, Stephanofilaria kaeli (Buckley Reference Buckley1937) in Malaysia, Stephanofilaria dedoesi (Buckley Reference Buckley1937) in Indonesia, and Stephanofilaria okinawaensis (Ueno et al. Reference Ueno, Chibana and Yamashiro1977; Ueno and Chibana Reference Ueno and Chibana1978) in Japan.
Stephanofilarial infections are well described in cattle. The key features of stephanofilarial infections in cattle include acanthosis, spongiosis, ulcers, eosinophilia, and fibrosis. Ulcerated lesions vary in size from 5 × 4 cm to 36 × 10 cm and are well defined, with crusted edges and a purulent exudate (Islam et al. Reference Islam, Azad, Akther, Sen, Avi and Juli2018; Loke and Ramachandran Reference Loke and Ramachandran1967; Matos et al. Reference Matos, Maldaner, Gruchouskei, Machado, Zuliani, Cavalca, Alves and Elias2022). In Australian cattle, the size of lesions was dependent on whether they were positive or not for stephanofilarian worms. Specifically, Stephanofilaria sp positive lesions were larger (ranging from 7.02 to 54.44 cm2) and had significantly higher scores of alopecia (>80% of the lesion area affected) and hyperkeratosis than lesions without Stephanofilarial worms (Naseem et al. Reference Naseem, Allavena, Raza, Constantinoiu, McGowan, Turni, Kamran, Tabor and James2023). Moreover, like previous studies, Stephanofilaria-infected lesions had significantly higher inflammation and higher scores of eosinophil, macrophage, and lymphocyte infiltration than lesions without Stephanofilaria worms (Naseem et al. Reference Naseem, Allavena, Raza, Constantinoiu, McGowan, Turni, Kamran, Tabor and James2023). In rhinoceros, lesions caused by Stephanofilaria dinniki are similar to Stephanofilarial lesions in cattle; they are erosive, crater-like dermal ulcerations, 2–3 cm deeper than the surrounding skin with crusting, and have edges raised above the normal skin (Tremlett Reference Tremlett1964). The average diameter of these lesions varies between 5–7 cm and 15–20 cm in black rhinoceros populations (Hitchins and Keep Reference Hitchins and Keep1970; Mutinda et al. Reference Mutinda, Otiende, Gakuya, Kariuki, Obanda, Ndeere, Ndambiri, Kariuki, Lekolool, Soriguer, Rossi and Alasaad2012; Plotz Reference Plotz2014; Tremlett Reference Tremlett1964). Although the prevalence of stephanofilarial ulcerative lesions is less in white compared to black rhinoceros, white rhinoceros appear to experience severe ulceration (King’ori et al. Reference King’ori, Waiguchu, Ruoro, Muriithi, Mumbi, Omondi, Aminga, Angwenyi, Mijele and Chiyo2024). In Meru National Park, the stephanofilarial lesions were 23 cm in diameter in white rhinoceros, and 15 cm in black rhinoceros (Mutinda et al. Reference Mutinda, Otiende, Gakuya, Kariuki, Obanda, Ndeere, Ndambiri, Kariuki, Lekolool, Soriguer, Rossi and Alasaad2012). Lesions in black rhinoceros are typically on the shoulder and ventral thorax, while in white rhinoceros, lesions are commonly seen at the rump (Mutinda et al. Reference Mutinda, Otiende, Gakuya, Kariuki, Obanda, Ndeere, Ndambiri, Kariuki, Lekolool, Soriguer, Rossi and Alasaad2012). Although these stephanofilarial infections are considered benign in black rhinoceros, a similar disease in cattle causes delayed puberty, reduced milk yields, and longer inter-calving intervals in affected cattle (Rai et al. Reference Rai, Srivastava, Sunder, Kundu and Jeykumar2010). Recent studies have shown that these hemorrhaging lesions are associated with a loss in body condition, anemia, elevated stress, and mortality in black rhinoceros (Mutinda et al. Reference Mutinda, Otiende, Gakuya, Kariuki, Obanda, Ndeere, Ndambiri, Kariuki, Lekolool, Soriguer, Rossi and Alasaad2012; Plotz Reference Plotz2014) and may be chronic or recrudescent (Kock and Kock Reference Kock and Kock1990). These hemorrhagic skin lesions can also cause black rhinoceros’ mortality through systemic secondary bacterial infection (Clausen and Ashford Reference Clausen and Ashford1980). Flaring up of wounds can also be induced by translocation-associated stress, particularly in artificially managed rhinoceros metapopulations where translocation is a common practice (Hitchins and Keep Reference Hitchins and Keep1970). Hemorrhaging lesions can also affect the aesthetic values of the rhinos.
Despite the economic importance of the genus Stephanofilaria to the livestock industry, molecular genetic studies and genetic identification have gained attention only recently (Lui et al. Reference Lui, Kulpa, Verocai, Armién, Edwards, Wiener and Rech2023; Naseem et al. Reference Naseem, Raza, Allavena, McGowan, Morgan, Constantinoiu, Tabor and James2021). However, no studies have focused on the genetic identification or molecular characterization of Stephanofilaria species infecting the critically endangered rhinoceros populations. Previous studies on this genus have focused on morphological features for species identification. However, diagnostic morphological features for species identification can be limited to specific life stages or sexes, and if these life stages or sexes are rare or absent or the key species diagnostic features of specimens are badly distorted, specimen identification can be impossible to achieve. Moreover, paucity of suitable keys remains a huge gap between the number of taxa treated in keys and the number of species for which gene sequence data are available (Will and Rubinoff Reference Will and Rubinoff2004). Molecular methods such as DNA-barcoding offer potentially efficient alternative approaches to species identification and studies of their ecology. This is because molecular techniques are sensitive in the detection of small quantities of nematode DNA, regardless of the developmental stage or sex. Molecular barcoding may help to identify cryptic and polymorphic species and give means to associate life history stages of unknown identity (Schander and Willassen Reference Schander and Willassen2005). Molecular genetic identification is also useful for the investigation of aspects of nematode biology, including understanding the life cycle and the identification of potential vectors. Genes used in molecular barcoding offer quick and reliable means of species identification and for detecting the transmission, spread, and the evolution of nematode species (Powers Reference Powers2004). Developing genetic data of pathogens can also be useful in inferring their transmission, reconstructing their epidemiological history, and identifying physical and environmental drivers of disease spread. Molecular methods such as DNA-barcoding beyond species detection can offer potentially efficient alternative approaches to studying the nematode disease ecology and evolution.
In this study, we isolated worms from ulcerative dermal lesion scrapings of black and white rhinoceros and conducted preliminary morphological characterization to corroborate previous descriptions of Stephanofilaria dinniki in rhinoceros. We also extracted DNA, carried out PCR amplification, and sequenced two gene fragments – the second internal transcribed spacer Ribosomal DNA (ITS-2) and the cytochrome c oxidase subunit 1 (Cox-1) genes – and used this for genetic identification and molecular characterization of Stephanofilaria dinniki for the first time. The second internal transcribed spacer (ITS-2) has a high degree of hyper-variability useful in discriminating populations of the same species, while the cytochrome c oxidase subunit 1 (Cox-1) gene is also useful in demonstrating finer intra-species level variation in many nematodes. Lastly, the patterns of sequence variation in the Cox-1 and ITS-2 were used to infer evolutionary and demographic signatures of the Stephanofilaria sp. infecting rhinoceros.
Materials and methods
Ethics statement
This study was approved by the Ethics Committee of the Kenya Wildlife Service (KWS/BRM/5001), the Institution mandated to protect and conserve Wildlife in Kenya. Stephanofilarial worms were collected from rhinoceros ulcerated skin lesions during immobilization of rhinoceros for ear notching, translocation, clinical management of injuries (due to snares and intraspecific fights), and clinical infections. Immobilization and translocation of rhinoceros are undertaken by experienced veterinarians guided by the Kenya Wildlife Service protocol for rhinoceros’ immobilization and translocation (https://www.kws.go.ke/file/3239/download?token=5F1MI56-) and guidelines on Wildlife Veterinary Practice 2018 and the Veterinary Surgeons and Veterinary Para-professionals Act Cap 366 of the Laws of Kenya that regulate veterinary practice in Kenya.
Study area
We collected worms from rhinoceros in two sanctuaries: Meru Rhino Sanctuary (MRS) and Ol Jogi Conservancy (OLJ) (Figure 1). MRS is a fenced portion in the western part of Meru National Park covering 38.8 km2 and is located between 36°40′ E–37°00′ E and 0°02′ S–0°07′ N. Meru National Park covers an area of 870 km2 and receives rainfall ranging from 635–762 mm in the west of the park to 305–356 mm in the east. The habitat of MNP varies from woodland to open grasslands intersected by permanent rivers and associated riverine vegetation. The southern portion of the rhino sanctuary is dominated by forests, while the rest of the sanctuary is dominated by thickets and grassland interspersed with several rivers such as Makutano, Kanjoo, and Rojaweru. Meru National Park has a rich diversity of wild animals including elephants, hippopotamuses, leopard, cheetah, black rhinoceros, and some rare antelopes, with incursions from cattle, camel, goats, and sheep. The Meru National Park is a home for 40 black rhinos and 79 white rhinos.

Figure 1. Map of Kenya showing rhinoceros sanctuaries and study sites (rhinoceros sanctuaries) sampled in this study.
Ol Jogi is on private land located in central Kenya between 37°00′E–37°05′E and 0°15′N–0020′N. Founded in 1960, it forms part of the Laikipia-Samburu-Meru-Marsabit ecosystem. It ranges between 1800 m and 1920 m in altitude and lies on the Laikipia plateau near Mount Kenya with an area of 235 km2 and provides a safe habitat for indigenous species. Rainfall averages 460 mm per year. The vegetation is a mosaic of grassland, Acacia woodland, and shrubs. In 2005, Ol Jogi expanded its 50-km2 rhino sanctuary to the entire conservancy and developed a fence, enabling rhinoceros to be protected within its boundary while allowing the free migration of all other species.
Isolation and morphological examination of worms
Skin scrapings from filarial wounds were taken from immobilized rhinos during routine notching exercises in MRS and OLJ. Filarial wounds were washed of mud, and a clean sterile scalpel blade was used for scraping (Figure 2). The scraped tissue was collected into a sterile fecal pot (Figure 2). Scrapings were collected from seven black rhinoceros in OLJ, four black rhinoceros from MRS, and three white rhinoceros from MRS during ear notching exercises. Ear notching was carried out in 2021 at MRS and in 2022 at OLJ. The scraping was done deep into the filarial wound crust until blood oozed. Later, the wound was dressed with a topical antibiotic spray. The scraping was then transferred to a 5-ml cryovial, labeled, and preserved in liquid nitrogen. Data on specific rhino identity, time of capture, date, and GPS coordinates were recorded for every scraped individual rhino. The samples were then transported to the laboratory for analysis. In the lab, frozen scraping samples were thawed and poured into a clean glass petri dish and examined using a Leica EZ4D stereo microscope to detect filarial worms. Detected worms were picked using a pair of pointed forceps, transferred on a clean glass slide, and observed for finer details on Leica DM 500 microscope. Following observation under a light microscope, each worm was then macerated using a clean scalpel blade and transferred to a 1-ml Eppendorf tube awaiting DNA extraction. Worms were successfully isolated in 7 of 11 black rhinoceros, but nothing was isolated from 3 white rhinoceros.

Figure 2. A photo showing a collection of scrapings from a filarial wound on a black rhino in Ol Jogi wildlife conservancy.
DNA extraction from the isolated worms
DNA was extracted from 25 worms from five rhinoceros sampled from OLJ, and from five worms extracted from two rhinoceros sampled from MRS. Each isolated worm was macerated and homogenized in 360 μl phosphate-buffered saline (PBS) (pH =7.4) and vortexed for 90 seconds. The extraction of total nucleic acids was done from 200 μl of the homogenate using DNeasy blood and tissue kit (QIAGEN, Hilden, Germany) following the manufacturer’s instructions. A total number of 35 worms were analyzed at the ITS-2 and Cox-1 loci. All the 35 worm samples were amplified at both loci and used for molecular identification of filaria worm. A 700 bp fragment of mitochondrial Cox-1 gene and a 900 bp of the ITS-2 gene-fragment were obtained using the primers sets: St_CO1F (5’- ATACTGTKAATCATAAGACTATTGG -3’) and St_CO1R: (5’- GACCAAAAAATCAAAACAAATGCTG -3’) (Naseem et al. Reference Naseem, Raza, Allavena, McGowan, Morgan, Constantinoiu, Tabor and James2021) and S_5.8S_29F: (5’- TAGCGGTGGATCACTTGGCTCG -3’) and 28S_400R: (5’- CAACTTTCCCTCACGGTACTTGT -3’) (Naseem et al. Reference Naseem, Raza, Allavena, McGowan, Morgan, Constantinoiu, Tabor and James2021). The amplification of the mitochondrial Cox-1 and the ITS-2 gene-fragments was carried out in a total volume of 25 μl that consisted of 1 μl DNA template, 12.5 μl of OneTaq® Quick-Load® 2X Master Mix with Standard Buffer (New England Biolabs-NEB, Massachusetts, USA), and 0.5 μl of 10 mM each forward and reverse primers. As a negative control, molecular grade nuclease-free water was used. The following cycling conditions were used for PCR amplification in a SimpliAmp thermal cycler (Life Technologies): an initial denaturation at 94°C for 1 minute followed by 35 cycles of denaturation at 94°C for 20 seconds, annealing at 46°C for 20 seconds, and 1 minute extension at 68°C. The final extension was at 68°C for 5 minutes.
The PCR amplicons were resolved in a 1% (W/V) agarose gel by electrophoresis with a 1x TAE running buffer at 90 V for 35 min. Gelpilot 1000 bp plus ladder (Qiagen, Germany) was used as a molecular size DNA marker. Ethidium Bromide-stained gels were visualized under UV trans-illumination. All the amplicons with the expected band size were submitted for sequencing in both forward and reverse directions at Macrogen Europe B.V. DNA chromatograms and sequences were visualized and edited using Geneious v11 (Kearse et al. Reference Kearse, Moir, Wilson, Stones-Havas, Cheung, Sturrock, Buxton, Cooper, Markowitz and Duran2012) software. ITS-2 sequences were manually edited for polymorphic or double chromatogram peaks. Consensus worm-sequences for ITS-2 and Cox-1 genes were generated from forward and reverse sequence data and exported as fasta files to DNAsp 6 (Rozas et al. Reference Rozas, Ferrer-Mata, Sánchez-DelBarrio, Guirao-Rico, Librado, Ramos-Onsins and Sánchez-Gracia2017) for diphasing and haplotype extraction, in the case of nuclear ITS-2. Clean edited sequences for the ITS-2 locus were obtained for 11 worms from three rhinoceros in OLJ and three worms from two rhinoceros sampled in MRS. For the Cox-1 locus, clean edited sequences were obtained for 19 worms from four rhinoceros in OLJ and for three worms from two rhinoceros in MRS. The remaining sequences with low quality chromatograms were discarded.
Genetic identification and phylogenetic analyses
The trimmed and edited DNA sequences were used to detect similarities with other available sequences in GenBank using BLASTn (https://blast.ncbi.nlm.nih.gov/Blast.cgi (accessed on 26 October 2023).
Phylogenetic analyses using Cox-1 and ITS-2 sequences of Stephanofilaria worms were carried out to identify the evolutionary clades they fall into using orthologous gene sequences of identified worms available from GenBank. The method of maximum likelihood and the best nucleotide substitution model estimated with ModelFinder (Kalyaanamoorthy et al. Reference Kalyaanamoorthy, Minh, Wong, von Haeseler and Jermiin2017) were used to infer evolutionary relationships between species using the IQTree software (Nguyen et al. Reference Nguyen, Schmidt, von Haeseler and Minh2014). Initial trees for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the best nucleotide substitution model for the data and then selecting the topology with a superior log-likelihood value. To evaluate statistical support for clades to which stephanofilarial worms were assigned to, the ultrafast bootstrap support approximation from 1,000 replicates using the UFBoot algorithm was used (Hoang et al. Reference Hoang, Chernomor, Von Haeseler, Minh and Vinh2018).
The Cox-1 maximum likelihood tree was fitted using the GTR+F+I+G4 nucleotide substitution model selected based on the Bayesian Information Criteria (BIC). The model had a rate heterogeneity of Gamma with four categories. The proportion of invariable sites was 0.423, and the Gamma shape parameter alpha was 0.808 for the Cox-1 locus. Twelve haplotypes obtained in this study (GenBank Accession numbers PP749201 — PP749212) and 32 orthologous sequences from known nematode species from GenBank, including two recent sequences of Stephanofilaria species, were used. The sequences consisted of 649 nucleotide sites with 325 (50.08% of all sites) constant sites and the 275 parsimony informative sites. The tree was rooted using Protospirura numidica as an out group. The robustness of the topology was tested using 1,000 bootstraps implemented with the UFBoot algorithm in the IQTree software program. The ITS-2 maximum likelihood tree was also fitted using a GTR+F+I+G4 substitution model selected based on the BIC. The model had a rate heterogeneity with four categories. The proportion of invariable sites was 0.334, and the gamma shape parameter alpha was 0.757. Eight haplotypes obtained in this study (GenBank Accession numbers PP892684 — PP892691) and 27 orthologous sequences from known nematode species including sequences of Stephanofilaria genus were used. The sequences consisted of 424 nucleotide sites with 212 (50%) constant sites and 171 (40.30%) parsimony informative sites. The tree was rooted using Stegophorus macronectes as an out group. The robustness of the topology was tested using 1,000 bootstraps implemented with the UFBoot algorithm in the IQTree software program.
Genetic diversity, natural selection, and demography
Genetic diversity indices including haplotype diversity, nucleotide diversity, number of haplotypes, and segregating sites were determined for the mitochondrial Cox-1 and the nuclear ITS-2 genes using DNASP 6.0 software (Rozas et al. Reference Rozas, Ferrer-Mata, Sánchez-DelBarrio, Guirao-Rico, Librado, Ramos-Onsins and Sánchez-Gracia2017). Haplotype diversity is the probability that two randomly chosen haplotypes are different (Nei and Roychoudhury Reference Nei and Roychoudhury1974; Nei and Tajima Reference Nei and Tajima1981), whereas nucleotide diversity is the average number of nucleotide differences per site, π, between any two randomly chosen DNA sequences from a population (Nei Reference Nei1987). Numerous tests were conducted to determine signatures of departures from neutral expectations and large and stable population sizes (Fu’s Fs and Tajima’s D, Fu and Li’s D*, Fu and Li’s F*, Ramos-Onsins and Rozas’s R2, Raggedness r) (Ramos-Onsins and Rozas Reference Ramos-Onsins and Rozas2002a) on the patterns of polymorphism of the Cox-1 and ITS-2 sequence data using DNASP 6.0 (Rozas et al. Reference Rozas, Ferrer-Mata, Sánchez-DelBarrio, Guirao-Rico, Librado, Ramos-Onsins and Sánchez-Gracia2017). Tajima D (Tajima Reference Tajima1989), Fu and Li’s D* and F* (Fu and Li Reference Fu and W-H1993) tests are sensitive to selection, whereas Fu’s FS (Fu Reference Fu1997), Ramos-Onsins and Rozas R2 (Ramos-Onsins and Rozas Reference Ramos-Onsins and Rozas2002a), and a raggedness index r (Harpending Reference Harpending1994) are sensitive to changes in demography (Ramos-Onsins and Rozas Reference Ramos-Onsins and Rozas2002a). Moreover, demographic changes are expected to affect the genome more evenly than selective pressures (Galtier et al. Reference Galtier, Depaulis and Barton2000; Hahn et al. Reference Hahn, Rausher and Cunningham2002), so analyses of the empirical distribution of Tajima’s D for the mitochondrial Cox-1 and nuclear ITS-2 regions could help decipher between selection and demography as explanations for the observed deviation from neutral expectation.
Results
Morphology of adult worms, larvae, and eggs
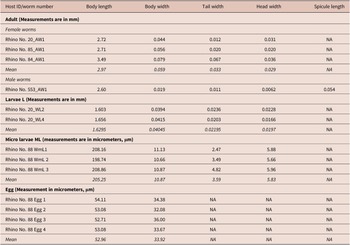
Several female worms and a single male were recovered from suspected filarial wound lesion scrapings. The single male recovered measured 2.6 mm in length and 0.02 mm in maximum thickness (Table 1). In addition, we observed a spicule on this male measuring 0.054 mm in length (Figure 3). Recovered adult female worms measured between 2.71 mm and 3.49 mm in length and 0.044 mm and 0.079 mm in maximum thickness (Table 1). Some females were observed with their uterine tubes containing either embryonated eggs or developed microfilaria (Figure 3). The eggs measured from 52 to 54 micrometers by 30 to 34 micrometers (Table 1). Microfilaria (L1) measured between 0.199 mm and 0.21 mm in length and 0.01 mm in diameter (Table 1). The body of the larvae (L1) is cylindrical in shape, the head blunt and the tail tapering to a point (Figure 3). Larvae, recovered from lesions likely at the L5 stage, measured 1.6 mm in length (Table 1, Figure 3). The cuticle throughout the length of the body of adult female worms was finely striated transversely (Figure 3). There was a lack of spines on the transverse striations and the oral opening was surrounded by numerous cuticularized spines (Figure 3). The head had a terminal peri-buccal ring of cuticular spines that we could not determine the number. Behind this ring is a circle of eight cephalic spines, arranged in pairs. The tails of adult females were prominently curved ventrally (Figure 3).
Table 1. Measurements of larva, adult, and egg dimension of Stephanofilaria sp isolated from black rhinoceros


Figure 3. Stephanofilaria dinniki showing key features, anterior portion (A), cleaned with glycerol (B), showing an oral opening surrounded by numerous cuticularized spines terminal peri-buccal ring of cuticular spines. The mid portion (C) shows fine transverse striations. Posterior portion (D), cleaned with glycerol (E), showing a curved tail, A male spicule is shown in (F), L5 Larvae in (G), a whole worm with a curved tail and L1 and embryonated eggs (I).
Genetic identification and phylogenetic analyses
Twenty-two worms from six rhinoceros were successfully amplified and sequenced at the Cox-1 locus comprising 649 bp after primer trimming, while 14 worms from six rhinoceros were amplified and sequenced at the ITS 2-locus comprising 424 sites resulting in 28 diphased sequences with 36 segregating sites.
Blast results were inconclusive due to lack of a barcoding gap between the taxa constituting the top four hits on GenBank based on BLASTn matching our orthologous sequences. Specifically, 10 of 12 Cox-1 haplotypes best matched Thelazia callipaeda with a similarity between 81% and 82.6% and sequence coverage of 71% to 97% followed by 9 of 12 haplotypes matching Serratospiculum tendo at a similarity of 81% and a sequence coverage of 97%. However, when orthologous sequences are matched against Stephanofilaria genus sequences on GenBank using BLASTn, all the 12 haplotypes sequences matched to Stephanofilaria stilesi isolated from American cattle with a percent similarity of 81.18% to 82.93% and a coverage of 51% to 62% to Stephanofilaria sp with a percent sequence identity of 79.04% to 80.69% and coverage of 93%. For the ITS-2 haplotypes, the best match identified by BLASTn using the GenBank database was Thelazia callipaeda isolate from a red fox with a 91.21% to 91.86% similarity and a sequence coverage of 75% and to Stephanofilaria sp isolated from Australian cattle by 88.66% to 89.00% similarity and sequence coverage of 71%. Other top similarity hits were Ochocerca gibsoni and Loloa loloa with a percentage identity of 85.29% to 85.67% and sequence coverage at about 83%.
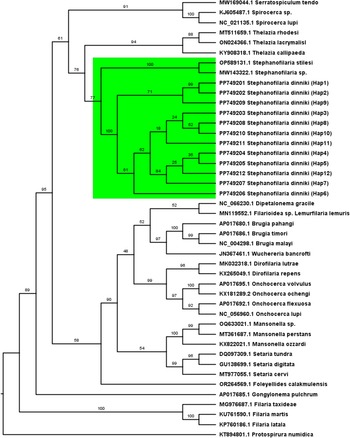
Phylogenetic analyses at the Cox-1 locus (Figure 4) and the ITS-2 locus (Figure 5) provided support that sequences from this study formed a single cluster. The cluster had 100% bootstrap support at the Cox-1 locus and 97% bootstrap support at the ITS-2 locus for belonging to the same cluster. Similarly, Stephanofilaria dinniki samples formed a cluster with other Stephanofilaria species. Specifically, there was 77% bootstrap support at the partial Cox-1 gene and a 93% support at the partial ITS-2 gene for a Stephanofilaria genus cluster.

Figure 4. Maximum likelihood tree based on partial Cytochrome c oxidase 1 gene showing a Stephanofilaria clade coloured green. Best-fit substitution model according to BIC: GTR+F+I+G4, Model of rate heterogeneity: Invariant + Gamma with 4 categories, Proportion of invariable sites = 0.423, Gamma shape alpha: 0.808. Bayesian information criterion (BIC) score: 15133.14. Input data: 44 sequences with 649 nucleotide sites; Number of constant sites: 325 (= 50.08% of all sites); Number of invariant (constant or ambiguous constant) sites: 325 (= 50.08% of all sites); Number of parsimony informative sites: 275, Number of distinct site patterns: 346.

Figure 5. Maximum likelihood tree based on partial ITS-2 gene showing a Stephanofilaria clade coloured green. Best-fit model according to BIC: GTR+F+I+G4. Model of rate heterogeneity: Invar + Gamma with 4 categories, Proportion of invariable sites: 0.3344, Gamma shape alpha: 0.757, Input data: 35 sequences with 424 nucleotide sites, Number of constant sites: 212 (50% of all sites), Number of parsimony informative sites: 171 (40.3% of all sites), Number of distinct site patterns: 244, Log-likelihood of the tree: -2961.660 (s.e. 125.580). Bayesian information criterion (BIC) score: 6389.149.
Genetic diversity, demography, and evolution
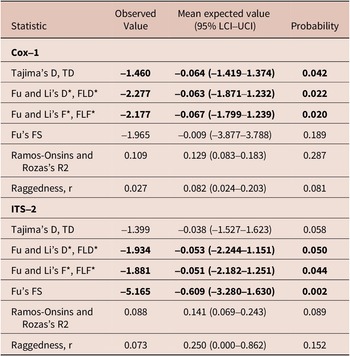
Twenty-two worms from six rhinoceros were successfully amplified and sequenced at the Cox-1 locus comprising 649 bp after primer trimming. About 54.5% of the 22 sequences were unique (i.e., 22 sequences comprised of twelve distinct haplotypes obtained) with 32 segregating sites. All the variations were synonymous substitutions. Haplotype diversity (Hd ± Sd) was high (0.930 ± 0.030), nucleotide diversity, PiT, was low (0.008 ± 0.002), and the average number of nucleotide differences, Kt, was 5.277 ± 2.650 (Table 2).
Table 2. Evolutionary and demographic signals on Cox-1 and ITS-2 for Stephanofilaria dinniki worms from Black rhinoceros
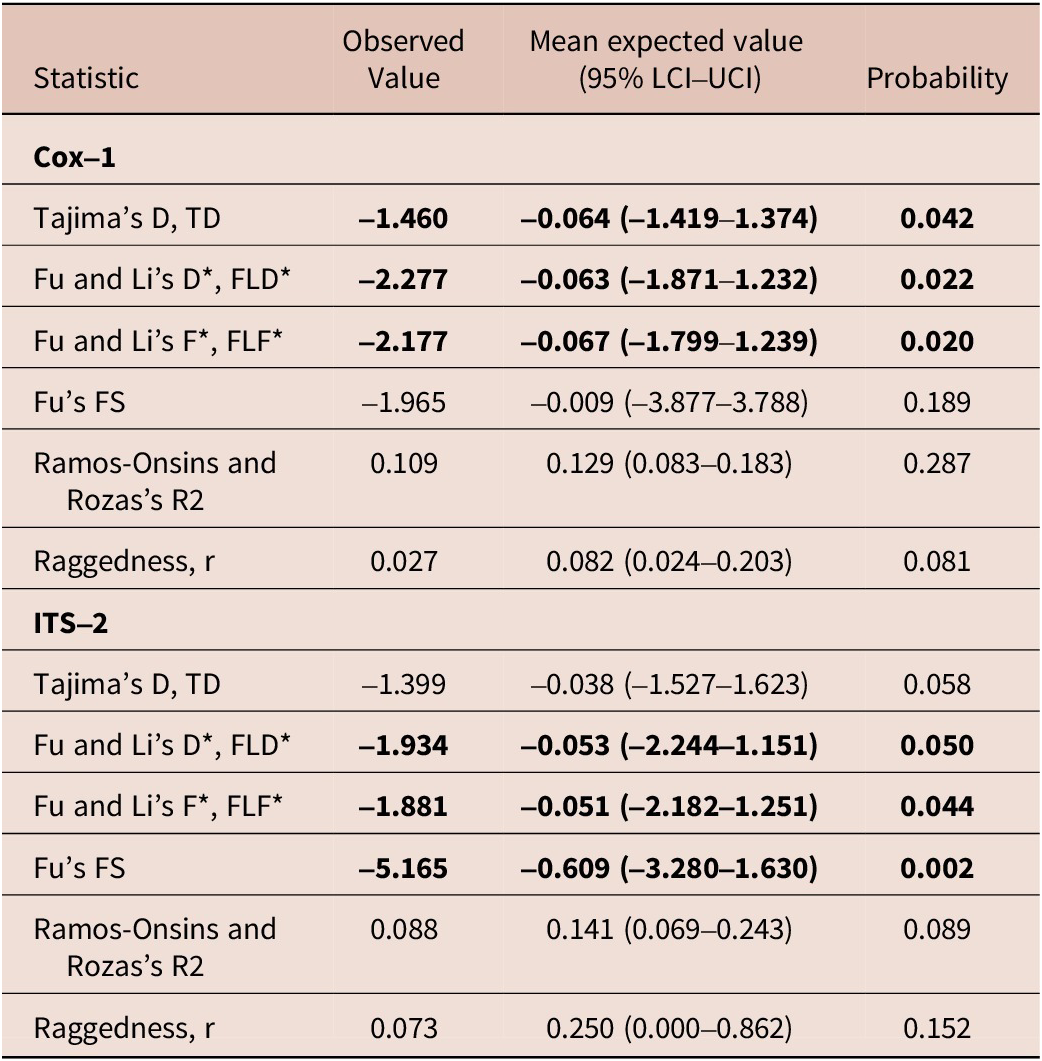
Fourteen worms from six rhinoceros were amplified and sequenced at the ITS-2 locus comprising 406 sites after trimming resulting in 28 diphased sequences with six segregating sites. There were eight distinct haplotypes (or 28.6% unique sequences) with Haplotype diversity, Hd ± Sd being 0.579 ± 0.104, nucleotide diversity, PiT, was 0.00197 ± 0.0016, and the average number of nucleotide differences, Kt, was 0.799 ± 0.595 (Table 2).
Tajima’s D and Fu and Li’s D* and F* for the parasite Cox-1 locus were negative and significantly different from neutral expectation and a large and stable population, but Fu’s Fs, Ramos-Onsins and Rozas’s R, and raggedness index r were not significantly different from neutral expectation. Similarly, neutrality tests such as Tajima’s D, TD, Fu and Li’s D*, FLD*, Fu and Li’s F*, FLF* and Fu’s FS were negative, but only Tajima’s D was not statistically significant from neutral expectation at the ITS-2 locus (Table 2).
Discussion
Morphological examination of worms recovered from black rhinoceros skin scrapings revealed the presence of diagnostic traits for the filaroid worm, Stephanofilaria dinniki. These traits include, among others, the possession of a cuticle along the entire length of the body with fine transverse striations, a cuticularized ring of numerous cuticular spines around the oral opening, and tails that are curved ventrally (Round Reference Round1964) in both males and females. In addition, female worms had uterine tubes containing either embryonated eggs or developed microfilaria (Hitchins and Keep Reference Hitchins and Keep1970). The generic diagnostic features for Stephanofilaria include presence of the oral aperture surrounded by a protruding cuticular rim with a denticulate edge (Bain et al. Reference Bain, Van der Lugt and Kazadi1996; Boomker et al. Reference Boomker, Bain, Chabaud and Kriek1995; Watrelot-Virieux and Pin Reference Watrelot-Virieux and Pin2006). Notable morphological deviation from previous studies was on size of the adult worms and larvae; the females from this study were smaller than in previous studies. For example, adults and micro larvae were 2.71–3.49 mm and 0.199–0.209 mm in this study compared to 4.6–5.7 mm and 0.12–0.15 mm from a previous study (Round Reference Round1964), respectively. Intraspecific size variation during various life stages has not been explicitly examined in this species, as only a single study has examined the body length variation in this species (Round Reference Round1964).
Phylogenetic analyses of the COX-1 and ITS-2 genes revealed a close relationship between Stephanofilaria worm sequences from this study and previously identified Stephanofilaria species. Using the Cox-1 sequences, stephanofilarial worms formed two sister clades: the Stephanofilaria dinniki clade and the Stephanofilaria stilesi clade (Stephanofilaria sp isolated from Australian cattle and Stephanofilaria stilesi isolated from American cattle) each with 100% bootstrap support. The two Stephanofilaria clusters formed a single clade with 77% bootstrap support. Similarly, phylogenetic analyses of the ITS-2 sequences also revealed that sequences of Stephanofilaria worms from rhinoceros clustered with known Stephanofilaria species with a 93% bootstrap support. Phylogenetic analyses at both the Cox-1 and ITS-2 loci suggest a closer relationship of the genus Stephanofilaria with Thelazioidea, rather than the family Filariidae (Filarioidea), in which it has been historically assigned to (Saparov et al. Reference Saparov, Akramova, Azimov and Golovanov2014). The close affinity to Thelazioidea is supported by recent genetic studies of this genus (Lui et al. Reference Lui, Kulpa, Verocai, Armién, Edwards, Wiener and Rech2023; Naseem et al. Reference Naseem, Raza, Allavena, McGowan, Morgan, Constantinoiu, Tabor and James2021).
For the Cox-1 gene, Tajima’s D, and Fu and Li’s D* and F* were negative and statistically significant, but the Fu’s FS, Fu and Li’s D*, and Fu and Li’s F*, but not Tajima’s D, were negative for the ITS-2 gene. Negative and significant values indicate an excess of rare polymorphisms, which suggests positive or purifying selection, genetic hitchhiking, and a recent spatial expansion or increase in population size. Statistical tests like Tajima’s D, Fu and Li’s D* and F*, and Fu’s FS can be significantly negative under purifying selection, population expansion, or selective sweeps, although each statistic may be best at detecting one of these forces (Braverman et al. Reference Braverman, Hudson, Kaplan, Langley and Stephad1995; Fu Reference Fu1998; Fu Reference Fu1995; Simonsen et al. Reference Simonsen, Churchill and Aquadro1995). Tajima’s D statistic and Fu and Li D* and F* tests are the most powerful in detecting a selective sweep and genetic hitchhiking (Simonsen et al. Reference Simonsen, Churchill and Aquadro1995). However, Fu’s FS, Ramos-Onsins and Rozas’s R2, and raggedness r tests are sensitive in detecting population growth (Ramos-Onsins and Rozas Reference Ramos-Onsins and Rozas2002b). The significance of Tajima’s D but not Fu’s FS at the Cox-1 locus suggests that this gene is under purifying selection. Demographic changes, however, would be expected to affect the genome more evenly than selection, particularly the ITS-2 genes, which are known to evolve neutrally subject to secondary structure constraints (Prahl et al. Reference Prahl, Khan and Deo2021). Indeed, the ITS-2 gene had a negative and statistically significant Fu’s FS statistic, indicative of a population expansion after a reduced ancestral effective population size (Avise Reference Avise2000).
Purifying or positive selection occurs when an allele is favored by natural selection. The frequency of the favored allele increases creating an excess of rare polymorphism from prior standing genetic variation in the population. Cytochrome c oxidase subunit I (Cox-1) is one of the major proteins responsible for oxidative phosphorylation (OXPHOS), a process that releases ATP in eukaryotes. It is usually subject to strong purifying selection in response to higher energy requirements or limited oxygen availability (Boratynski et al. Reference Boratynski, Melo-Ferreira, Alves, Berto, Koskela, Pentikainen, Tarroso, Ylilauri and Mappes2014; Shen et al. Reference Shen, Liang, Zhu, Zhou, Irwin and Zhang2010; Tomasco and Lessa Reference Tomasco and Lessa2011) driven usually by altitudinal changes in temperature and associated hypoxia. Such environmental pressures have been suggested to cause changes in the structure and function of proteins associated with oxidative phosphorylation in mitochondria including the Cox-1, CytB, and the NADH dehydrogenase complex (Bartáková et al. Reference Bartáková, Bryjová, Nicolas, Lavrenchenko and Bryja2021). A similar evolutionary pattern for mitochondrial OXPHOS genes has been observed in deep sea fishes and subterranean mammals (Shen et al. Reference Shen, Pu, Chen, Murphy and Shen2019; Tomasco and Lessa Reference Tomasco and Lessa2011), vertebrates in environments that have total darkness, cold, scarce food, and low oxygen. Selected sweeps can occur as adaptation to new environments and habitats (Wei et al. Reference Wei, Silva-Arias and Tellier2023). A recent study revealed that Stephanofilaria infections in the Kenyan rhinoceros population is higher in sanctuaries with low minimum temperatures compared to the 1960s when it was more widespread including lowland sanctuaries with higher minimum temperature (King’ori et al. Reference King’ori, Waiguchu, Ruoro, Muriithi, Mumbi, Omondi, Aminga, Angwenyi, Mijele and Chiyo2024). The more recent restricted distribution of Stephanofilaria is possibly due to the extinction of Rhinomusca brucei Malloch (Parsons and Sheldrick Reference Parsons and Sheldrick1964), a biting fly previously associated with skin lesions caused by Stephanofilaria dinniki (Round Reference Round1964) in lowland areas (Mihok et al. Reference Mihok, Moloo, Oden’y, Brett, Rakwar, Munyoki, Kiilu and Kyorku1996). This extinction occurred due to reduction of rhinoceros numbers and the amalgamation of all remnant rhinoceros populations into a few populations in cooler highland locations. Moreover, Rhinomusca dutoiti, a species of fly that appears adopted to cooler temperatures in arid and semi-arid locations of other rhinoceros reserves, may have taken advantage of increased rhino densities to proliferate and become the dominant vector for Stephanofilaria dinniki.
Genetic diversity in this study was high (0.930 ± 0.030) for the Cox-1 gene and moderate (0.579 ± 0.104) for the ITS-2 gene. Nucleotide diversity was generally low, at 0.008 ± 0.002 for the Cox-1 gene and 0.00197 ± 0.0016 for the ITS-2 gene. The Cox-1 gene has been shown to display wider genetic variation among nematode species and similar patterns of diversity recorded in this study have been observed elsewhere. In the mulberry root-knot nematode, Meloidogyne enterolobii in China, haplotype diversity of 0.90 (Shao et al. Reference Shao, Zhang, You, Li, Feng and Xie2020) was observed for the Cox-1 gene. Similarly, the Cox-1 gene of the pinniped hookworms Uncinaria lucasi was reported to have high haplotype diversity ranging between 0.96 and 0.98 and a high nucleotide diversity at 0.014 (Davies et al. Reference Davies, Pagan and Nadler2020). A study of Haemonchus placei and Haemonchus contortus also revealed high haplotypic diversity (0.98–0.99 and 0.98–1) and nucleotide diversity (0.016–036, 0.009–0.01) at the Cox-1 locus, respectively, from several locations in Brazil (Brasil et al. Reference Brasil, Nunes, Bastianetto, Drummond, Carvalho, Leite, Molento and Oliveira2012). In contrast, several studies have also revealed moderate to low gene diversity at the Cox-1 gene. For example, in a study of Ascaris spp infecting humans, moderate diversity (0.616) was recorded in selected municipalities in Brazil (Monteiro et al. Reference Monteiro, Calegar, Santos, Bacelar, Coronato-Nunes, Reis, Boia, Carvalho-Costa and Jaeger2019). In another study on Heterotakis gallinarum infecting chicken in Tunisia, low genetic diversity at the Cox-1 gene ranging from 0.12 to 0.42 was observed at different locations (Amor et al. Reference Amor, Farjallah, Mohammed, Alagaili and Bahri-Sfar2018). Similar levels of nucleotide diversity at the Cox-1 gene (0.006 + 0.0006) have been observed in Thelazia callipaeda from China (Zhang et al. Reference Zhang, Shi, Han, Xiong, Yi, Jiang, Wang, Shen, Cui and Wang2018). Lower levels of nucleotide diversity were observed at the Cox-1 gene (0.00147 ± 0.00051) for Setaria digitata, a nematode causing ocular lesions in horses (Junsiri et al. Reference Junsiri, Kamkong, Chinkangsadarn, Ouisuwan and Taweethavonsawat2023). The ITS-2 gene also displays greater variability in gene and nucleotide diversity among nematodes. Gene diversity of the ITS-2 gene of Onchocerca volvulus in Africa and Brazil was 0.983, and nucleotide diversity was 0.0073 (Morales-Hojas et al. Reference Morales-Hojas, Cheke and Post2007). The mean gene diversity in 12 Haemonchus contortus populations from Thailand was 0.724 ± 0.025, while average nucleotide diversity was 0.007 ± 0.004 (Mangkit et al. Reference Mangkit, Thaenkham, Adisakwattana, Watthanakulpanich, Jantasuriyarat and Komalamisra2014). A study on Heterotakis gallinarum infecting chickens in Tunisia revealed low to moderate gene diversity ranging from 0.35 to 0.59 at the ITS genes (Amor et al. Reference Amor, Farjallah, Mohammed, Alagaili and Bahri-Sfar2018). The high gene diversity and low nucleotide diversity (Hd: 0.930 ± 0.030, PiT: 0.008 ± 0.002) at the Cox-1 observed in this study indicate a high number of closely related haplotypes, and suggest that the parasite population may have undergone a recent expansion following a bottleneck (Mendez-Harclerode et al. Reference Mendez-Harclerode, Strauss, Fulhorst, Milazzo, Ruthven and Bradley2007; Zhang et al. Reference Zhang, Dan, Wang, Liu, Zhou, Ma, Ren, Fu, Geng, Luo, Xie, Peng and Zhong2021). The parasite population likely expanded following the amalgamation of rhinoceros to cooler highland sanctuaries, an increase in rhinoceros population size and proliferation of rhinoceros sanctuaries in highland locations. These results are also consistent with a soft selective sweep as discussed earlier.
This study provides the first genetic characterization of Stephanofilaria dinniki and sheds light into the molecular diagnosis of Stephanofilaria dinniki in rhinoceros. Accurate identification of rhinoceros Stephanofilaria is crucial for its diagnosis, treatment, epidemiological studies, and control. This study also corroborates recent genetic studies, which suggested that this genus is phylogenetically closer to Thelazioidea rather than the family Filariidae (Filarioidea), where it is currently placed.
Data availability statement
All the relevant data has been deposited in Genbank or included in the main manuscript.
Acknowledgements
We grateful to the Kenya Wildlife Service Management for funding management activities that generated data used in this study.
Competing interest
The coauthor and I have no conflict of interest regarding the work we have done, the science or with the donor(s).